The goal of splinetrials is to analyze longitudinal clinical data applying natural cubic splines to a continuous time variable.
Installation
Install this package from CRAN with:
install.packages("splinetrials")You can also install the development version of splinetrials from GitHub with:
# install.packages("pak")
pak::pak("NikKrieger/splinetrials")Methods
The NCS model is a longitudinal mixed model of repeated measures (MMRM) with random effects marginalized out, additive baseline covariates, and time parameterized using natural cubic splines. The parameterization has additive effects and pairwise interactions for study arm and time point (i.e., spline basis functions). The model is constrained so that all study arms have equal means at baseline.
Data
We begin with a clinical data set with one row per patient per scheduled visit (including baseline). The following simulated data set represents the general structure (e.g. column names) but not necessarily realistic responses.
set.seed(1)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
data <-
mutate(
cross_join(
data.frame(
patient = do.call(paste0, expand.grid(LETTERS, letters))[1:50],
arm =
sample(c("control", "active1", "active2"), 50, replace = TRUE),
categorical1 = sample(paste0("level", 1:3), 50, replace = TRUE),
categorical2 = sample(c("level1", "level2"), 50, replace = TRUE),
continuous1 = rnorm(50),
continuous2 = rnorm(50)
),
data.frame(
time_observed_index = seq_len(8),
time_scheduled_index = seq_len(8),
time_scheduled_label =
c("Baseline", paste("visit", c(3, 4, 5, 7, 8, 9, 12))),
time_scheduled_continuous = seq(from = 0, length.out = 8) / 4
)
),
time_observed_continuous =
time_scheduled_continuous + runif(n(), min = -0.1, max = 0.1),
response =
rnorm(
n(),
10 - (1 * (arm == "active1") + 2 * (arm == "active2") +
3 * (arm == "control")) * sqrt((time_observed_index / 4) -
min(time_observed_index / 4))
)
)
head(data)
#> patient arm categorical1 categorical2 continuous1 continuous2
#> 1 Aa control level3 level2 0.6466744 -0.9106807
#> 2 Aa control level3 level2 0.6466744 -0.9106807
#> 3 Aa control level3 level2 0.6466744 -0.9106807
#> 4 Aa control level3 level2 0.6466744 -0.9106807
#> 5 Aa control level3 level2 0.6466744 -0.9106807
#> 6 Aa control level3 level2 0.6466744 -0.9106807
#> time_observed_index time_scheduled_index time_scheduled_label
#> 1 1 1 Baseline
#> 2 2 2 visit 3
#> 3 3 3 visit 4
#> 4 4 4 visit 5
#> 5 5 5 visit 7
#> 6 6 6 visit 8
#> time_scheduled_continuous time_observed_continuous response
#> 1 0.00 0.07296151 10.685015
#> 2 0.25 0.18543891 11.766415
#> 3 0.50 0.49866375 8.439280
#> 4 0.75 0.73594267 7.332906
#> 5 1.00 1.01285277 6.027557
#> 6 1.25 1.28123246 6.099311The following is a summary of the required columns in the data set. Here are the columns not related to time points or visits:
-
response: User-supplied numeric vector of responses to the clinical endpoint. Could beAVAL. -
patient: User-supplied character vector, ID of the patient in the study. UsuallyUSUBJID. -
arm: User-supplied character vector, study arm of the given patient. -
categorical1,categorical2: User-supplied example categorical baseline covariates. -
continuous1,continuous2: User-supplied example continuous baseline covariates.
Here are the columns related to time points or visits:
-
time_observed_continuous: User-supplied numeric vector of continuous times corresponding to each observed patient visit. Varies from patient to patient. Units could be in years, months, or days, or some other unit. Must be on the same time scale astime_scheduled_continuous. (If the former is in weeks, then the latter must also be in weeks.) -
time_observed_index: User-supplied integer index vector of bins of observed time points. These numeric labels should indicate groupings of time points based on roundingtime_observed_continuousto a small set of strategic fixed time points. -
time_observed_factor: Derived ordered factor version oftime_observed_indexto structure the correlation matrix. Derived asordered(as.character(time_observed_index), levels = as.character(sort(unique(time_observed_index)))). -
time_scheduled_index: User-supplied integer index vector of scheduled visit numbers. UsuallyAVISITN. -
time_scheduled_label: User-supplied character vector of human-readable scheduled visit labels. UsuallyAVISIT. -
time_scheduled_continuous: User-supplied numeric vector of continuous times of when each visit was scheduled. It must be on the same time scale astime_scheduled_continuous, the values must have a one-to-one correspondence with the scheduled visits intime_scheduled_label, and the baseline must have a value of 0. If the time scale is weeks, then the study team can create this variable in advance usingtime_scheduled_label(e.g.AVISIT). For example, ifAVISITis"Baseline", thentime_scheduled_continuousmust be 0. Iftime_scheduled_labelis"Visit Number 5 (Week 24)", thentime_scheduled_continuousshould be 24.
Main analysis
The NCS analysis is a call to a single function: ncs_analysis(). This produces a table of summary statistics, including LS means and confidence intervals. All arguments to ncs_analysis() should be exposed as user inputs.
library(splinetrials)
results_table <- ncs_analysis(
data = data,
response = "response",
subject = "patient",
arm = "arm",
control_group = "control",
time_observed_continuous = "time_observed_continuous",
time_observed_index = "time_observed_index",
time_scheduled_continuous = "time_scheduled_continuous",
time_scheduled_label = "time_scheduled_label",
covariates = ~ continuous1 + categorical2,
cov_structs = c("us", "toeph", "ar1h", "csh", "cs"),
df = 3
)
results_table
#> # A tibble: 24 × 32
#> arm time n est sd se lower upper response_est response_se
#> <fct> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 active1 Baseline 19 10.0 1.04 0.240 9.57 10.5 9.95 0.127
#> 2 active1 visit 3 19 9.13 0.909 0.208 8.72 9.54 9.40 0.105
#> 3 active1 visit 4 19 9.02 1.10 0.253 8.53 9.52 8.94 0.144
#> 4 active1 visit 5 19 9.08 0.969 0.222 8.65 9.52 8.69 0.138
#> 5 active1 visit 7 19 8.57 1.17 0.268 8.05 9.10 8.63 0.134
#> 6 active1 visit 8 19 8.72 0.861 0.198 8.34 9.11 8.70 0.150
#> 7 active1 visit 9 19 9.16 1.21 0.278 8.61 9.70 8.84 0.126
#> 8 active1 visit 12 19 8.83 0.851 0.195 8.45 9.21 9.03 0.183
#> 9 active2 Baseline 13 10.3 0.883 0.245 9.82 10.8 9.95 0.127
#> 10 active2 visit 3 13 8.49 1.10 0.304 7.90 9.09 9.19 0.120
#> # ℹ 14 more rows
#> # ℹ 22 more variables: response_df <dbl>, response_lower <dbl>,
#> # response_upper <dbl>, change_est <dbl>, change_se <dbl>, change_df <dbl>,
#> # change_lower <dbl>, change_upper <dbl>, change_test_statistic <dbl>,
#> # change_p_value <dbl>, diff_est <dbl>, diff_se <dbl>, diff_df <dbl>,
#> # diff_lower <dbl>, diff_upper <dbl>, diff_test_statistic <dbl>,
#> # diff_p_value <dbl>, percent_slowing_est <dbl>, …The table has one row per study arm per timepoint of interest and the following columns.
-
arm: study arm, i.e. treatment group -
time: discrete visit time labels from thetime_scheduled_labelcolumn in the data. -
n: number of non-missing observations. -
est: observed mean response in the data. -
sd: observed standard deviation of the response in the data. -
se: observed standard error of the response in the data (justse / sqrt(n)). -
lower: lower bound of an observed 95% confidence interval in the data. -
upper: upper bound of an observed 95% confidence interval in the data. -
response_est: LS mean of the response. -
response_se: standard error of the LS mean of the response. -
response_df: degrees of freedom of the LS mean of the response. -
response_lower: lower 95% confidence bound of the LS mean of the response. -
response_upper: upper 95% confidence bound of the LS mean of the response. -
change_est,change_lower,change_upper,change_se,change_df: like the analogousresponse_*columns, but for LS mean change from baseline. -
change_test_statistic: test statistic of a two-sided test that the LS mean change from baseline is not equal to 0, at a significance level of 5%. -
change_p_value: same aschange_test_statistic, but the p-value instead of the test statistic. -
diff_est,diff_se,diff_df,diff_lower,diff_upper,diff_test_statistic,diff_p_value: same as the analogouschange_*columns, but for treatment differences. -
percent_slowing_estLS mean of percent slowing (on the percentage scale). -
percent_slowing_lower,percent_slowing_upper: a conservative approximation to a confidence interval on percent slowing. Assumes uncorrelated changes from baseline. -
correlation: correlation structure of the whole model. -
optimizer: optimization algorithm for the whole model.
We can plot the model against the data.
ncs_plot_means(results_table)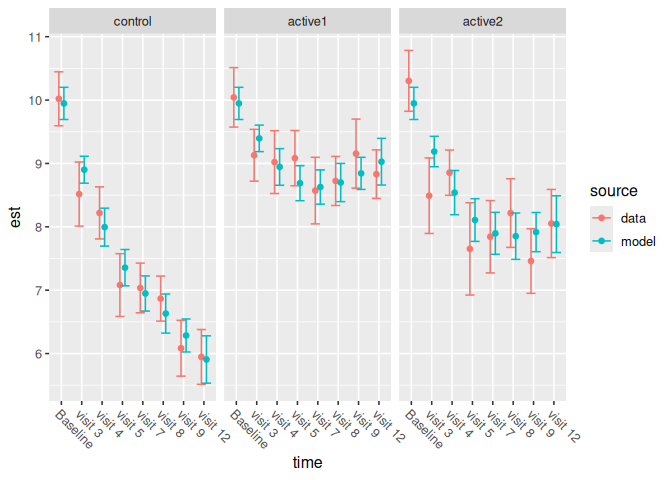
Subgroup analysis
The subgroup analysis is similar, but it begins with a data set with a categorical variable column to indicate subgroup membership.
set.seed(1)
data_with_subgroup <-
mutate(
cross_join(
data.frame(
patient = do.call(paste0, expand.grid(LETTERS, letters))[1:120],
arm = rep(c("control", "active1", "active2"), each = 40),
subgroup = rep(c("subgroup1", "subgroup2", "subgroup3"), times = 40),
categorical1 = sample(paste0("level", 1:3), 120, replace = TRUE),
categorical2 = sample(c("level1", "level2"), 120, replace = TRUE),
continuous1 = rnorm(120),
continuous2 = rnorm(120)
),
data.frame(
time_observed_index = seq_len(8),
time_scheduled_index = seq_len(8),
time_scheduled_label =
c("Baseline", paste("visit", c(3, 4, 5, 7, 8, 9, 12))),
time_scheduled_continuous = as.numeric(seq(from = 0, length.out = 8) / 4)
)
),
time_observed_continuous =
time_scheduled_continuous + runif(n(), min = -0.1, max = 0.1),
response = rnorm(
n(),
mean =
10 - (1 * (arm == "active1") + 2 * (arm == "active2") +
3 * (arm == "control")) * sqrt((time_observed_index / 4) -
min(time_observed_index / 4))
)
)
head(data_with_subgroup)
#> patient arm subgroup categorical1 categorical2 continuous1 continuous2
#> 1 Aa control subgroup1 level1 level1 -0.9568919 0.6060734
#> 2 Aa control subgroup1 level1 level1 -0.9568919 0.6060734
#> 3 Aa control subgroup1 level1 level1 -0.9568919 0.6060734
#> 4 Aa control subgroup1 level1 level1 -0.9568919 0.6060734
#> 5 Aa control subgroup1 level1 level1 -0.9568919 0.6060734
#> 6 Aa control subgroup1 level1 level1 -0.9568919 0.6060734
#> time_observed_index time_scheduled_index time_scheduled_label
#> 1 1 1 Baseline
#> 2 2 2 visit 3
#> 3 3 3 visit 4
#> 4 4 4 visit 5
#> 5 5 5 visit 7
#> 6 6 6 visit 8
#> time_scheduled_continuous time_observed_continuous response
#> 1 0.00 -0.07428706 9.336270
#> 2 0.25 0.23840196 9.916441
#> 3 0.50 0.43845954 6.507177
#> 4 0.75 0.73698012 7.510785
#> 5 1.00 0.94502668 6.533178
#> 6 1.25 1.34221926 9.607641For completeness, here is a full description of the columns in the data. Columns not related to time points or visits:
-
subgroup: User-supplied character vector indicating the subgroup of each patient. -
response: User-supplied numeric vector of responses to the clinical endpoint. Could beAVAL. -
patient: User-supplied character vector, ID of the patient in the study. UsuallyUSUBJID. -
arm: User-supplied character vector, study arm of the given patient. -
categorical1,categorical2: User-supplied example categorical baseline covariates. -
continuous1,continuous2: User-supplied example continuous baseline covariates.
Columns related to time points and visits:
-
time_observed_continuous: User-supplied numeric vector of continuous times corresponding to each observed patient visit. Varies from patient to patient. Units could be in years, months, or days, or some other unit. Must be on the same time scale astime_scheduled_continuous. (If the former is in weeks, then the latter must also be in weeks.) -
time_observed_index: User-supplied integer index vector of bins of observed time points. These numeric labels should indicate groupings of time points based on roundingtime_observed_continuousto a small set of strategic fixed time points. -
time_observed_factor: Derived ordered factor version oftime_observed_indexto structure the correlation matrix. Derived asordered(as.character(time_observed_index), levels = as.character(sort(unique(time_observed_index)))). -
time_scheduled_index: User-supplied integer index vector of scheduled visit numbers. UsuallyAVISITN. -
time_scheduled_label: User-supplied character vector of human-readable scheduled visit labels. UsuallyAVISIT. -
time_scheduled_continuous: User-supplied numeric vector of continuous times of when each visit was scheduled. It must be on the same time scale astime_scheduled_continuous, the values must have a one-to-one correspondence with the scheduled visits intime_scheduled_label, and the baseline must have a value of 0. If the time scale is weeks, then the study team can create this variable in advance usingtime_scheduled_label(e.g.AVISIT). For example, ifAVISITis"Baseline", thentime_scheduled_continuousmust be 0. Iftime_scheduled_labelis"Visit Number 5 (Week 24)", thentime_scheduled_continuousshould be 24.
The NCS subgroup analysis is a call to a single function: ncs_analysis_subgroup(). This produces a table of summary statistics, including LS means and confidence intervals.
subgroup_results <-
ncs_analysis_subgroup(
data = data_with_subgroup,
response = "response",
subject = "patient",
arm = "arm",
control_group = "control",
subgroup = "subgroup",
subgroup_comparator = "subgroup1",
time_observed_continuous = "time_observed_continuous",
time_observed_index = "time_observed_index",
time_scheduled_continuous = "time_scheduled_continuous",
time_scheduled_label = "time_scheduled_label",
covariates = ~ continuous1 + categorical2,
cov_structs = c("us", "toeph", "ar1h", "csh", "cs"),
df = 3
)We can plot the model against the data.
ncs_plot_means_subgroup(subgroup_results$within)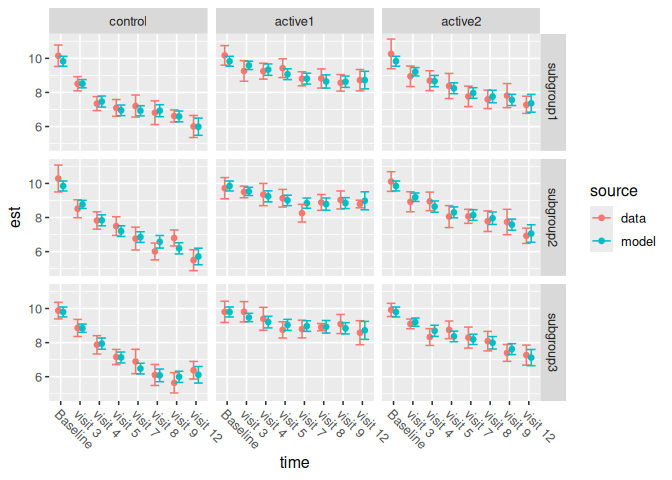
Between-subgroup table:
subgroup_results$between
#> # A tibble: 72 × 30
#> arm time subgroup n est sd se lower upper response_est
#> <fct> <chr> <fct> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 active1 Baseline subgroup1 13 10.2 1.05 0.292 9.60 10.7 9.83
#> 2 active1 Baseline subgroup2 14 9.72 1.19 0.317 9.10 10.3 9.85
#> 3 active1 Baseline subgroup3 13 9.81 1.14 0.317 9.18 10.4 9.80
#> 4 active2 Baseline subgroup1 13 10.3 1.60 0.444 9.39 11.1 9.83
#> 5 active2 Baseline subgroup2 13 10.1 1.06 0.295 9.53 10.7 9.85
#> 6 active2 Baseline subgroup3 14 9.92 0.741 0.198 9.53 10.3 9.80
#> 7 control Baseline subgroup1 14 10.2 1.20 0.320 9.52 10.8 9.83
#> 8 control Baseline subgroup2 13 10.3 1.45 0.402 9.50 11.1 9.85
#> 9 control Baseline subgroup3 13 9.88 0.896 0.249 9.39 10.4 9.80
#> 10 active1 visit 3 subgroup1 13 9.26 1.11 0.307 8.66 9.86 9.59
#> # ℹ 62 more rows
#> # ℹ 20 more variables: response_se <dbl>, response_df <dbl>,
#> # response_lower <dbl>, response_upper <dbl>, change_est <dbl>,
#> # change_se <dbl>, change_df <dbl>, change_lower <dbl>, change_upper <dbl>,
#> # change_test_statistic <dbl>, change_p_value <dbl>, diff_subgroup_est <dbl>,
#> # diff_subgroup_se <dbl>, diff_subgroup_df <dbl>, diff_subgroup_lower <dbl>,
#> # diff_subgroup_upper <dbl>, diff_subgroup_test_statistic <dbl>, …Within-subgroup table:
subgroup_results$within
#> # A tibble: 72 × 33
#> arm time subgroup n est sd se lower upper response_est
#> <fct> <chr> <fct> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 active1 Baseline subgroup1 13 10.2 1.05 0.292 9.60 10.7 9.83
#> 2 active1 visit 3 subgroup1 13 9.26 1.11 0.307 8.66 9.86 9.59
#> 3 active1 visit 4 subgroup1 13 9.25 0.856 0.237 8.78 9.71 9.34
#> 4 active1 visit 5 subgroup1 13 9.42 1.01 0.280 8.87 9.97 9.07
#> 5 active1 visit 7 subgroup1 13 8.80 0.762 0.211 8.39 9.22 8.81
#> 6 active1 visit 8 subgroup1 13 8.82 1.03 0.286 8.26 9.38 8.64
#> 7 active1 visit 9 subgroup1 13 8.56 0.893 0.248 8.08 9.05 8.63
#> 8 active1 visit 12 subgroup1 13 8.72 1.15 0.319 8.09 9.34 8.72
#> 9 active2 Baseline subgroup1 13 10.3 1.60 0.444 9.39 11.1 9.83
#> 10 active2 visit 3 subgroup1 13 8.95 1.10 0.306 8.35 9.55 9.21
#> # ℹ 62 more rows
#> # ℹ 23 more variables: response_se <dbl>, response_df <dbl>,
#> # response_lower <dbl>, response_upper <dbl>, change_est <dbl>,
#> # change_se <dbl>, change_df <dbl>, change_lower <dbl>, change_upper <dbl>,
#> # change_test_statistic <dbl>, change_p_value <dbl>, diff_arm_est <dbl>,
#> # diff_arm_se <dbl>, diff_arm_df <dbl>, diff_arm_lower <dbl>,
#> # diff_arm_upper <dbl>, diff_arm_test_statistic <dbl>, …The between-subgroup and within-subgroup tables have one row per study arm per timepoint per subgroup and the following columns.
-
arm: study arm, i.e. treatment group -
time: discrete visit time labels from thetime_scheduled_labelcolumn in the data. -
subgroup: subgroup level -
n: number of non-missing observations. -
est: observed mean response in the data. -
sd: observed standard deviation of the response in the data. -
se: observed standard error of the response in the data (justse / sqrt(n)). -
lower: lower bound of an observed 95% confidence interval in the data. -
upper: upper bound of an observed 95% confidence interval in the data. -
response_est: LS mean of the response. -
response_se: standard error of the LS mean of the response. -
response_df: degrees of freedom of the LS mean of the response. -
response_lower: lower 95% confidence bound of the LS mean of the response. -
response_upper: upper 95% confidence bound of the LS mean of the response. -
change_est,change_lower,change_upper,change_se,change_df: like the analogousresponse_*columns, but for LS mean change from baseline. -
change_test_statistic: test statistic of a two-sided test that the LS mean change from baseline is not equal to 0, at a significance level of 5%. -
change_p_value: same aschange_test_statistic, but the p-value instead of the test statistic. -
diff_arm_est,diff_arm_se,diff_arm_df,diff_arm_lower,diff_arm_upper,diff_arm_test_statistic,diff_arm_p_value: same as the analogouschange_*columns, but for treatment differences. -
diff_subgroup_est,diff_subgroup_se,diff_subgroup_df,diff_subgroup_lower,diff_subgroup_upper,diff_subgroup_test_statistic,diff_subgroup_p_value: same as the analogous*_arm_*columns, but for the subgroup differences. -
percent_slowing_estLS mean of percent slowing (on the percentage scale). -
percent_slowing_lower,percent_slowing_upper: a conservative approximation to a confidence interval on percent slowing. Assumes uncorrelated changes from baseline. -
correlation: correlation structure of the analysis subgroup model. -
optimizer: optimizer of the analysis subgroup model.
Type-III ANOVA fixed effects:
subgroup_results$type3
#> # A tibble: 15 × 6
#> effect chisquare_test_stati…¹ df p_value correlation optimizer
#> <chr> <dbl> <int> <dbl> <chr> <chr>
#> 1 spline_fn(time_o… 134. 1 4.36e-31 heterogene… mmrm+tmb
#> 2 spline_fn(time_o… 342. 1 2.23e-76 heterogene… mmrm+tmb
#> 3 spline_fn(time_o… 189. 1 6.16e-43 heterogene… mmrm+tmb
#> 4 subgroup 0.0641 2 9.68e- 1 heterogene… mmrm+tmb
#> 5 continuous1 1.72 1 1.90e- 1 heterogene… mmrm+tmb
#> 6 categorical2 0.958 1 3.28e- 1 heterogene… mmrm+tmb
#> 7 spline_fn(time_o… 0.770 2 6.80e- 1 heterogene… mmrm+tmb
#> 8 spline_fn(time_o… 0.189 2 9.10e- 1 heterogene… mmrm+tmb
#> 9 spline_fn(time_o… 0.514 2 7.73e- 1 heterogene… mmrm+tmb
#> 10 spline_fn(time_o… 24.7 2 4.26e- 6 heterogene… mmrm+tmb
#> 11 spline_fn(time_o… 376. 2 2.02e-82 heterogene… mmrm+tmb
#> 12 spline_fn(time_o… 42.0 2 7.62e-10 heterogene… mmrm+tmb
#> 13 spline_fn(time_o… 13.9 4 7.74e- 3 heterogene… mmrm+tmb
#> 14 spline_fn(time_o… 2.62 4 6.23e- 1 heterogene… mmrm+tmb
#> 15 spline_fn(time_o… 2.54 4 6.37e- 1 heterogene… mmrm+tmb
#> # ℹ abbreviated name: ¹chisquare_test_statisticThere is also a table for the subgroup interaction test:
subgroup_results$interaction
#> model aic bic loglik -2*log(l) test_statistic df
#> 1 reduced model 2884.131 3040.231 -1386.066 2772.131 NA NA
#> 2 full model 2891.246 3080.796 -1377.623 2755.246 16.88529 12
#> p_value correlation optimizer
#> 1 NA heterogeneous unstructured mmrm+tmb
#> 2 0.1539634 heterogeneous unstructured mmrm+tmbThe test itself is a likelihood ratio test between a full model with subgroup-treatment interaction and a reduced model without subgroup-treatment interaction. The table has a row for the full model, a row for the reduced model, and the following columns.
-
model: the model, either full or reduced. -
df: degrees of freedom -
aic: Akaike Information Criterion -
bic: Bayesian Information Criterion -
loglik: log likelihood -
test_statistic: test statistic of the likelihood ratio test. -
p_value: p-value of the likelihood ratio test. -
correlation: correlation structures of the full and reduced models of the interaction test. -
optimizer: optimizers of the full and reduced models of the interaction test.